THE CURRENT PARADIGM APPLIED TO PLANTS: A MODEL OF THE PLANT OSCILLATOR
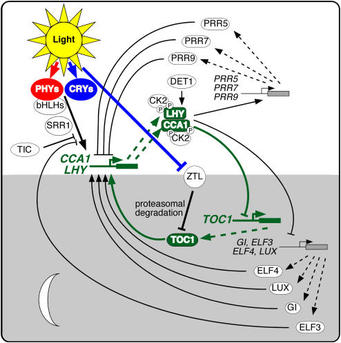
The sequencing of the Arabidopsis genome (Arabidopsis Genome Initiative, 2000) identified no obvious orthologs to most known clock proteins, which means that theArabidopsis clock mechanism is novel, at least in terms of its composition. Nonetheless, the paradigm of interlocked feedback loops seems to be conserved. A number of recent reviews discuss the increasingly complex picture of theArabidopsis clock (Salomé and McClung, 2004, 2005b; Harmon et al., 2005; Mizuno and Nakamichi, 2005). A simplified version of the Arabidopsis circadian clock is illustrated in Figure 3 (see also Table 1). It comprises three interlocked feedback loops, with two single Myb domain transcription factors, CIRCADIAN AND CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), playing roles in each loop. TOC1, the founding member of a family of pseudo-response regulators (PRRs), closes one loop, while three TOC1 paralogs, PRR5, PRR7, and PRR9, close a second loop. A third loop includes a Myb transcription factor, LUX ARRHYTHMO (LUX).
How do we conclude this? A toc1 loss-of-function mutant was identified as a short period mutant through a forward genetic screen, as described above. If the oscillating mRNA and protein abundance of a clock component, such as TOC1, is necessary for oscillator function but becomes pegged at a constant high level through overexpression, arrhythmicity should result. Indeed, TOC1 overexpressors are arrhythmic (Makino et al., 2002; Más et al., 2003b). CCA1 was identified initially as binding to the LHCB1*3 promoter. Loss of CCA1 function causes short period (Green and Tobin, 1999), but its overexpression causes arrhythmicity (Wang and Tobin, 1998), suggesting that it too is a core clock component. LHY, CCA1's closest paralog in Arabidopsis, was identified in a screen for late-flowering mutants. The allele identified in a screen of transposon-tagged mutants turned out to be overexpressed, which conferred arrhythmicity (Schaffer et al., 1998; Wang and Tobin, 1998). lhy loss of function, like that of cca1, confers short period, but the cca1 lhy double mutant is arrhythmic, suggesting that they are core clock components that function redundantly (Alabadí et al., 2002; Mizoguchi et al., 2002).How these genes form an oscillator loop is not completely understood. CCA1 and LHY bind to the TOC1 promoter, and overexpression of either results in low levels ofTOC1 expression, consistent with their roles as negative regulators of TOC1. TOC1 is inferred to be a positive regulator because expression of CCA1 and LHY is greatly reduced in a severe toc1-2 mutant (Alabadí et al., 2001, 2002; Matsushika et al., 2002b; Mizoguchi et al., 2002; Harmer and Kay, 2005; Hazen et al., 2005). AlthoughTOC1 overexpression results in arrhythmicity, neither CCA1 nor LHY expression levels are dramatically elevated (Makino et al., 2002). TOC1 contains a CCT (for CONSTANS, CONSTANS-LIKE, TOC1) domain thought to be involved in transcription (Strayer et al., 2000) but has not been shown to bind to either CCA1 orLHY promoters. It seems that TOC1 on its own is insufficient for expression of CCA1 and LHY. Several other genes, including GIGANTEA (GI), EARLY FLOWERING3(ELF3), ELF4, and LUX, are required for CCA1 and LHY expression (Park et al., 1999; Doyle et al., 2002; Mizoguchi et al., 2002; Hazen et al., 2005).In other systems, the oscillator has been shown to include multiple interlocked feedback loops. Consistent with this paradigm, modeling studies show that available data cannot be accounted for within a single feedback loop (Locke et al., 2005). At least two other loops are thought to interlock with the TOC1/CCA1/LHY loop. Locke et al. (2005) proposed a second loop in which TOC1 is activated by a hypothetical evening-expressed protein that itself is repressed by TOC1 and demonstrated thatGI behavior was consistent with that predicted for this hypothetical component. A number of investigators have proposed a third loop. CCA1 and LHY are positive regulators of three TOC1 relatives, PRR5, PRR7, and PRR9 (Farré et al., 2005;Harmer and Kay, 2005; Mizuno and Nakamichi, 2005). PRR5/7/9 are negative regulators of CCA1/LHY because CCA1 and LHY transcripts accumulate in prr7 andprr7 prr9 mutants (Farré et al., 2005), and CCA1 is constitutively transcribed in the arrhythmic prr5 prr7 prr9 triple mutant (Nakamichi et al., 2005b). PRR5/7/9 and TOC1 are thought to be mutually repressive (Mizuno and Nakamichi, 2005). Loss of function of prr7 or prr9 causes period lengthening, while loss of function of prr5causes period shortening (Kaczorowski and Quail, 2003; Michael et al., 2003). The circadian phenotypes of the single prr mutants are small (period changes of 1 to 1.5 h) compared with the period shortening (3 to 4 h) seen in toc1-2 mutants, but redundancy among the PRRs may partially account for this. The phenotype of theprr7 prr9 double mutant is more than additive; the period lengthening is dramatically increased, and the double mutant is arrhythmic in the dark (Farré et al., 2005;Nakamichi et al., 2005a; Salomé and McClung, 2005a). Emphasizing the centrality of the PRRs to clock function, the triple prr5 prr7 prr9 mutant is essentially arrhythmic under all conditions tested (Nakamichi et al., 2005b). However, overexpression of PRR3, PRR5, or PRR9 has only small period effects (Matsushika et al., 2002a; Sato et al., 2002; Murakami et al., 2004), suggesting that additional factors are required for full PRR function.Transcriptional regulation is important in clock function, but it is clear that posttranscriptional regulation is an essential constituent of the clock mechanism. While incompletely understood, casein kinase II (CK2) phosphorylates CCA1 and LHY, and overexpression of the regulatory β3 subunit (CKB3) confers short period (Sugano et al., 1998, 1999). CK2-mediated phosphorylation of CCA1 is necessary for in vivo function (Daniel et al., 2004). LHY is degraded via the proteasome, and this is accelerated in det1-1, providing a molecular explanation of the period-shortening effect of this mutation (Song and Carré, 2005), although a role for phosphorylation in degradation remains possible. Recently, a second type of posttranslational modification has been implicated in clock function. SPINDLY is an N-acetylglucosamine transferase that decorates GI, among other targets, and spymutants exhibit altered rhythms in leaf movement (Tseng et al., 2004).The identification of a novel family of proteins, ZEITLUPE (ZTL), LOV KELCH PROTEIN2 (LKP2), and FLAVIN binding KELCH REPEAT F-BOX (FKF), with PAS/LOV domains, Kelch repeats, and F-boxes (Nelson et al., 2000; Somers et al., 2000; Schultz et al., 2001), has contributed to our understanding of the role of protein degradation in the Arabidopsis circadian clock. The LOV domains are similar to those of phototropins, and the LOV domain of FKF is photoactive (Imaizumi et al., 2003). FKF seems restricted to photoperiodism (Nelson et al., 2000), but ZTL andLKP2 affect the clock (Somers et al., 2000; Jarillo et al., 2001; Schultz et al., 2001).ztl-1 and ztl-2 mutants are affected in the period length of numerous rhythms (Somers et al., 2000, 2004; Dodd et al., 2004). ZTL mRNA abundance is not clock regulated, but ZTL protein levels peak around dusk, while trough levels are reached around dawn (Kim et al., 2003). The rate of proteasome-mediated degradation of ZTL varies during the course of the day: ZTL is more stable at dusk, around its peak value, and is more rapidly degraded at dawn when it reaches its trough. F-box proteins provide specificity to proteasomal degradation pathways by specific interaction with and polyubiquitination of targets for degradation. In this case, ZTL is a component of an SCF complex that recruits TOC1 for proteasomal degradation (Somers et al., 2000; Más et al., 2003a; Han et al., 2004). In the ztl mutant, protein levels of TOC1 are elevated and only weakly rhythmic, demonstrating that ZTL is critical for degradation of TOC1. Increasing expression of ZTL confers corresponding dosage-dependent period shortening (Han et al., 2004). Collectively, these data argue that the level of TOC1 activity, as regulated through transcriptional repression by CCA1 and LHY and via protein degradation by ZTL, is a key determinant of circadian period.
